Junk DNA: A Journey Through the Dark Matter of the Genome (34 page)
Read Junk DNA: A Journey Through the Dark Matter of the Genome Online
Authors: Nessa Carey

Figure 17.2
The words shown in
bold and underlined
should do the trick for you: one of the most romantic and seductive first lines of poetry in the English language, ‘Had we but world enough and time’, from Andrew Marvell’s ‘To His Coy Mistress’.
In any long stretch of random letters there will also be combinations that spell words just by chance. Use these words by mistake when wooing (does anyone still woo?) the object of your desires and you may ruin your one chance for happiness. Figure 17.3 will show you how.
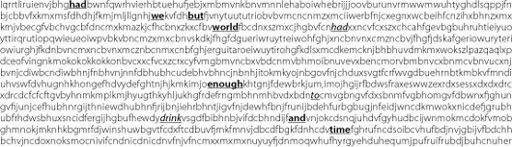
Figure 17.3
No! Bad combination! With a selection of
right
and
wrong
words, the sentiment may be very different, e.g. ‘Had we but had enough to drink’.
By using this slightly bizarre example, we can understand some of the mechanistic challenges that our cells face when splicing RNA molecules properly. If we were designing this as a process, it would have the components shown in Figure 17.4.
3
In addition to the components described in this diagram, it’s important to realise that different cells will handle the same gene differently, depending on the cell type and what is happening to it at any given moment. Consequently, all the stages have to be appropriately regulated and integrated so that the correct protein variants are made to meet the needs of the situation.

Figure 17.4
The sequence, reading from the top, lays out the steps that the splicing machinery has to be able to carry out to join up the appropriate amino acid-coding regions to create the correct mature messenger RNA.
The splice of life
This splicing of long RNAs to create smaller messenger RNAs that carry the information for specific proteins is a really complex process. It’s a very ancient system, and the components and steps have been maintained from yeast throughout the entire animal kingdom. It is carried out by a huge conglomeration of molecules called the spliceosome, which forms the splicing machinery. The spliceosome is composed of hundreds of proteins and also some junk RNAs, a little like the ribosomes that act as the factories to produce proteins.
4
One of the critical stages is that the spliceosome wraps around the intervening sequences that need to be removed from an RNA molecule. It snips them out and then joins up the amino acid-coding regions. It’s an enormously complicated multi-stage process but we know that one of the first key steps is that the spliceosome needs to recognise the intervening regions, so that it can bind to them and remove them.
The beginnings and ends of these intervening sequences are always indicated by particular two-base sequences. Junk RNA molecules in the spliceosome can bind to these two-base sequences in much the same way as the two strands of DNA can pair up in our genes.
But there are only four bases in RNA, which means there are only sixteen two-base sequences (AC and CA are considered as different pairs, as are all the others). We would expect that the two-base sequences that mark the beginnings and ends of the intervening sequences would also be found elsewhere in these sequences, and also in the amino acid-coding regions. This is indeed the case. So although these two-base sequences are necessary for splicing, they aren’t sufficient on their own to direct the process properly. Other sequences are also required, as indicated in Figure 17.5.
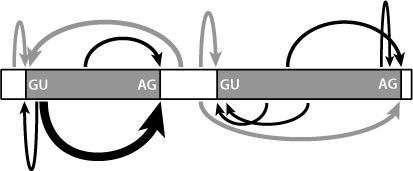
Figure 17.5
Multiple sequences within an RNA molecule interact to drive splicing. The two-base motifs shown are necessary but not in themselves sufficient to regulate all the fine-tuning of this process. Other sites are involved, of varying strengths, as indicated by the different sizes of arrows.
The other sequences involved in selecting how splicing will take place are found in both the junk intervening regions and the amino acid-coding regions. Some of them influence splicing very strongly, others are more subtle. Some increase the chances of a splice event, others decrease them. They work in complex partnerships and the impact that they have on the final splicing pattern is affected by other things happening in the cell, such as the precise complement of proteins in the spliceosome. The descriptions that are used for these modifying sequences usually include such words as ‘dizzying’ or ‘bewildering’. These are geek speak for ‘unbelievably complicated, way beyond anything we can get our heads around or even design predictive computer algorithms for at the moment.’
Splicing and disease
We can get clues to the degree of sophistication by looking at a group of genetic diseases. These include a form of blindness called retinitis pigmentosa, which affects about one in 4,000 people. The blindness is progressive, often starting in the teenage years with a decline in night vision, and then becoming steadily worse and more disabling with age. The loss of vision occurs because the cells in the eye that detect light gradually die off.
5
About one in twenty
cases is caused by a mutation in one of five proteins involved in a specific step in splicing.
6
,
7
,
8
,
9
The mutation only causes a deficit in the cells of the retina, and not in all the other cells in the body which also rely on splicing. This shows us that splicing is under complex cell- and gene-specific control, in ways that we haven’t yet been able to understand.
By contrast, there is a very severe form of dwarfism with other unusual features such as dry skin, sparse hair, seizures and learning disabilities. Affected children almost always die before they are four years old.
10
It’s very rare except in the Ohio Amish community, where 8 per cent of the people are carriers. That’s because the mutation that causes this condition was present in the small number of families that founded this community. It isn’t found in other Amish groups such as those in Pennsylvania, which were founded by other families. When the mutation that causes this condition was identified, the researchers first thought that it was changing the amino acid sequence of a gene that codes for a splicing protein. But we now know that the change actually disrupts the three-dimensional structure of a junk RNA that forms part of the spliceosome.
11
Unlike the retinitis pigmentosa situation, this defect in the action of the spliceosome causes a very wide-ranging set of symptoms, possibly by causing mis-splicing of lots of different genes.
Human disorders don’t just occur because of defects in the splicing machinery. They can also arise because protein-coding genes themselves have mutations in sites that are important for the control of splicing of the RNA from that single gene. Some authors have claimed that up to 10 per cent of human inherited disorders may be caused by mutations at the splice sites, those two-base sequences shown in Figure 17.5.
12
One example of this mechanism was a family in which two young siblings developed intractable diarrhoea within a few days of birth. Medical staff managed to stabilise the children, but the
diarrhoea persisted for many months and one of the two affected children died at seventeen months of age. When the genomes of the children were sequenced, the researchers found a mutation in a splice site in a gene, changing one of the GU sequences shown in Figure 17.5. This resulted in the splicing machinery skipping over an amino acid-coding region inappropriately. Essentially, an amino acid-coding region was left out of the protein, and as a consequence the protein could no longer do its job.
13
Kaposi’s sarcoma is a cancer that first came to public attention when it was found at high levels in people with AIDS. AIDS is caused by the human immunodeficiency virus (HIV) and the effect of the HIV infection is to suppress the immune system. Kaposi’s sarcoma is caused by a different virus called HHV-8. Normally our immune systems control this virus but if the immune system is seriously below par, HHV-8 can become established and trigger Kaposi’s sarcoma.
HHV-8 is present in a high percentage of people in the Mediterranean basin, but Kaposi’s sarcoma is rare in this population, and almost never found in small children. So medics were very surprised when a Turkish family brought in their two-year-old daughter who had a classic lesion characteristic of this cancer on her lip. The cancer spread rapidly and aggressively and the little girl died just four months after she was first diagnosed.
The child was negative in all tests for HIV. Her parents were related to each other, a first-cousin marriage. Researchers looked for genetic reasons why the daughter might have an impaired immune response to HHV-8.
By sequencing DNA obtained from samples that had been taken from the deceased girl, scientists identified a mutation in a splice site of a specific gene. The mutation changed an AG to an AA, which meant the spliceosome could no longer recognise where it was meant to cut the RNA molecule. The result was that a junk region that should have been removed was retained in the
messenger RNA molecule. This messed up the sequence, creating a stop signal much too early in the messenger RNA. This prevented the ribosome from making the full-length protein. Because the protein is one that is required for mounting a good immune response to viruses such as HHV-8, the child with the mutation was very susceptible to Kaposi’s sarcoma.
14
Although splice site mutations are relatively common, genetic diseases are more often caused by mutations in the amino acid-coding regions of genes. Some of these cause problems because they introduce stop signals that prevent the ribosomes from making full-length proteins from messenger RNA templates. Other mutations may change the code from one amino acid to another. For example, CAC codes for the amino acid histidine whereas CAG codes for glutamine, a different amino acid. But researchers have speculated that up to 25 per cent of the mutations that change the amino acid in this way also influence the splicing of nearby regions in the messenger RNA. In some cases the disease may be due not to the single amino acid alteration per se, but to the variation that the nucleotide change creates in the way a messenger RNA is spliced.
The problem is that it is very difficult to demonstrate that this is the case in most situations. Even if we can show that the change in the RNA leads to both an altered splicing pattern and an amino acid change, how can we tell which effect causes the disease symptoms? Are these due to protein with one altered amino acid, or because the protein has also been spliced in an unusual pattern?
Nature has actually provided us with proof that sometimes a mutation in a coding region can cause a disease by influencing splicing, rather than by changing an amino acid. There is an extraordinary disorder called Hutchinson-Gilford Progeria, named after the two scientists who first identified it.
Progeria
means early ageing and this particular form is incredibly dramatic. It is also extremely rare, affecting about one in 4 million children.
15
