Read Junk DNA: A Journey Through the Dark Matter of the Genome Online
Authors: Nessa Carey
Junk DNA: A Journey Through the Dark Matter of the Genome (14 page)

This would normally seem like quite a good bet for explaining why this female patient had developed this disorder. There was only one problem. The patient had a sister. A twin sister. An identical twin sister, derived from the very same egg and sperm. And her twin sister was absolutely healthy. No symptoms of Duchenne muscular dystrophy at all. How on earth could two women who were genetically absolutely identical differ so much with respect to a genetically inherited disorder?
Think back to those hundred or so cells that undergo X inactivation during early embryonic development. Just by chance, about 50 per cent of them will switch off one X chromosome, and the remainder will switch off the other one. The same pattern of X inactivation is passed on to all the daughter cells throughout life.
The sister with Duchenne muscular dystrophy was simply incredibly unlucky during this stage. Just by sheer chance, all the cells that would ultimately give rise to muscle switched off the normal copy of the X chromosome. This was the one inherited from her father. This meant that the only X chromosome switched on in her muscle cells was the faulty one from her carrier mother. So none of the affected twin’s muscle cells were able to express dystrophin and she developed the symptoms normally only seen in males.
When her genetically identical twin was developing, however, some of the cells that would give rise to muscle switched off the
normal X chromosome and some switched off the mutated one. This meant that her muscles expressed enough dystrophin to keep them healthy, and she was an asymptomatic carrier, just like her mother.
17
It is quite extraordinary to think that this was all caused by a simple fluctuation in the distribution of Xist, a long bit of RNA derived from junk DNA. The fluctuation lasted no more than a couple of hours, and occurred over a distance considerably less than one-millionth of the diameter of a human hair. Yet it was the difference between winning and losing in the health lottery.
Luck can be patchy
It is perhaps even stranger to think that some of the cat lovers among us look at, and stroke, the consequences of X inactivation every day. Tortoiseshell or calico cats (depending on which side of the Atlantic you’re reading this book) are the ones with the distinctive patterns of orange and black. These coat colours occur in patches. The gene that controls the coat colour comes in two forms. An individual X chromosome carries either the orange version or the black version.
If the X chromosome carrying the black version is inactivated, the orange version on the other chromosome will be expressed and vice versa. When the cat embryo is at the size of a hundred cells or so, one or other X chromosome will be inactivated in each cell. And just as in all the other examples, all the daughter cells will switch off the same X chromosome. Eventually, some of these daughter cells will give rise to the cells that create pigment in the fur. As more and more of these cells divide and develop, they stay close to each other. This means that daughter cells tend to be clustered in patches. Because of the pattern of X inactivation in the daughter cells, this will lead to patches of orange fur and patches of black fur. This process is shown in Figure 7.2.
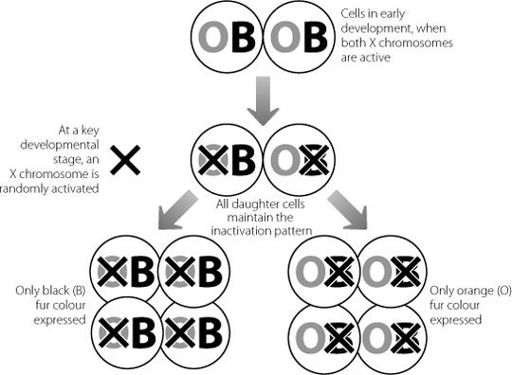
Figure 7.2
Schematic showing how patches of orange or black fur develop in female tortoiseshell cats depending on random X chromosome inactivation. The genes for fur colour lie on the X chromosome. If the black version is on the chromosome that is inactivated in a cell during early development, all descendants of that cell will only express the orange gene. The situation is reversed if the X chromosome carrying the orange gene is inactivated.
In 2002 scientists demonstrated beautifully just how random the process of X inactivation really is, by cloning a calico cat. They took cells from an adult female cat, and carried out the standard (but still fiendishly tricky) process of cloning. To do this, they removed the nucleus from the adult cat cell and put it into a cat egg whose own chromosomes they’d removed. This egg was implanted into a surrogate cat mother, and a lively and beautiful female kitten was born. And she didn’t look anything like the genetically identical cat of which she was a clone.
18
When this procedure is used to clone animals, the egg treats the new nucleus as if it was the real product of an egg fusing with
a sperm. It strips off as much information as possible from the DNA, taking it back to its basic genetic sequence. This doesn’t happen as effectively as in a real egg and sperm, which is one of the reasons why the success rate of this type of cloning is still very low. But sometimes it does work, as was the case here, and a cloned animal is born.
When the nucleus from the mother cat was put inside a cat egg, the egg caused changes to the chromosomes. One of these changes was the removal of the inactivating proteins on one of the X chromosomes, and the switching off of Xist expression. So for a short period in early development, both copies of the X chromosome were active. As the embryo developed, it went through the normal process at around the 100-cell stage of randomly inactivating an X chromosome in each cell. The pattern of X inactivation was passed on to daughter cells in the standard way, and the kitten thereby developed a different distribution of orange and black fur from its clonal ‘parent’.
The moral of this story? If you have a calico cat you think is exceptionally beautiful, take lots of videos, lots of photos and if you want to be very weird about it, call in a taxidermist when she dies. But if you are ever approached by a door-to-door travelling cloner, just send them on their way.
Footnotes
a
This gene is called
SRY
.
b
The name Xist is derived from X-inactive (Xi)-specific transcript.
c
Bases rather than base pairs, because RNA is single-stranded.
d
The gene is called MeCP2 and its role is to bind to epigenetically modified (methylated) DNA, where it interacts with other proteins and represses gene expression at the sites where it binds.
8. Playing the Long Game
For quite a few years, Xist was considered an anomaly, a strange molecular outlier with an extraordinarily unusual impact on gene expression. Even when Tsix was identified, it was possible to think that junk RNAs were restricted to the vital but unique process of X inactivation. It is only in recent years that we have begun to recognise that the human genome expresses thousands of this type of molecule, and that they are surprisingly important in normal cellular function.
We now categorise Xist and Tsix as members of a large class known as the long non-coding RNAs. The term is a somewhat misleading one, because of course what it means is non-coding with respect to proteins. As we shall see, the long non-coding RNAs do code for functional molecules. The functional molecules are the long non-coding RNAs themselves.
Long non-coding RNAs are defined rather arbitrarily as molecules which are greater than 200 bases in length, and which don’t code for proteins, making them different from messenger RNA. 200 bases is the lower size limit, but the biggest long non-coding RNAs can be 100,000 bases in length. There are lots of them, although no agreement yet on the precise number. Estimates range from 10,000 to 32,000 in the human genome.
1
,
2
,
3
,
4
But although there are a lot of long non-coding RNAs, they don’t tend to be expressed to as high a level as the classical messenger RNAs which code for proteins. Normally, the expression level of a long
non-coding RNA is less than 10 per cent of the level of an average messenger RNA.
5
This relatively low abundance of any one long non-coding RNA is one of the reasons why we have tended to disregard this type of molecule until fairly recently. Essentially, when the expression of RNA molecules from cells was analysed, the long non-coding RNAs simply could not be detected very reliably because the technology wasn’t sensitive enough. However, now that we know about their existence, we might think we should be able to analyse the genome of any organism, including humans, and predict their existence from the DNA sequence. We are, after all, pretty good at doing that for protein-coding genes.
But there are a number of aspects that make this difficult. We can identify putative protein-coding genes because of a number of features. They have certain sequences near the beginning and end of the genes that help us to find them. They also encode predicted runs of amino acids, which again give us confidence that a protein-coding gene may be present. Finally, most protein-coding genes are pretty similar if you look at a specific gene in different species. This means that if we identify a classical gene in an animal such as a pufferfish, it’s easy to use that sequence as a basis for analysing the human genome to see if we can predict the presence of a similar gene in ourselves.
However, long non-coding RNAs don’t have such strong sequence indicators as protein-coding genes, and they are also poorly conserved across species. Consequently, knowing the sequence of a long non-coding RNA in another species may not help us to identify a functionally related sequence in the human genome. Less than 6 per cent of a specific class of long non-coding RNAs in zebrafish, a common model system, have clearly equivalent sequences in mice and humans.
6
Only about 12 per cent of the same class of long non-coding RNAs that are found in humans and mice can be detected elsewhere in the animal kingdom.
7
,
8
The relatively poor conservation of long non-coding RNAs was confirmed in a recent study comparing expressed long non-coding RNAs from various tissues of different tetrapod species. Tetrapod refers to all land-living vertebrates along with those that have ‘returned to the sea’ such as whales and dolphins. This paper reported that there were 11,000 long non-coding RNAs that were only found in primates. Only 2,500 were conserved across tetrapods, of which a mere 400 were classified as ancient, by which the authors meant that they had originated over 300 million years ago, around the time when amphibians and other tetrapods diverged. The authors suspected that the ancient long non-coding RNAs are the ones that are most actively regulated in all organisms, and are probably mostly involved in early development.
9
Most vertebrates look very similar during the earliest stages of embryogenesis, so it may make sense that we and all our distant cousins are using similar pathways to get started.
The generally poor conservation across species has led some authors to speculate that the long non-coding RNAs are not very important. The rationale behind this is that if they were significant they would be more constrained to remain similar during evolution and the development of species; whereas instead, the sequences coding for these ‘junk’ RNAs are evolving much more rapidly than the ones that encode proteins.
Although this is a fair point, it’s perhaps an over-simplification. Long non-coding RNA molecules may be long in terms of the number of bases they contain, but that doesn’t necessarily mean they are elongated stringy molecules in the cell. This is because long RNA molecules can fold onto themselves, forming three-dimensional structures. The bases in RNA pair up, following similar rules to the way in which the two strands of DNA are bonded together. RNA is a single-stranded molecule, so its bases pair up over relatively short distances, bending the molecule into complex stable shapes. These 3D structures may be very important
in the function of the long non-coding RNA, and it’s possible that the 3D structure is conserved across species, even if the base sequence is not.
10
This is shown in Figure 8.1. Unfortunately, predicting similar structures is difficult to do using sequence data, limiting the usefulness of this technique in helping us to find functionally conserved long non-coding RNAs.