Connectome (25 page)
Authors: Sebastian Seung

Â
 Â
Â
Â
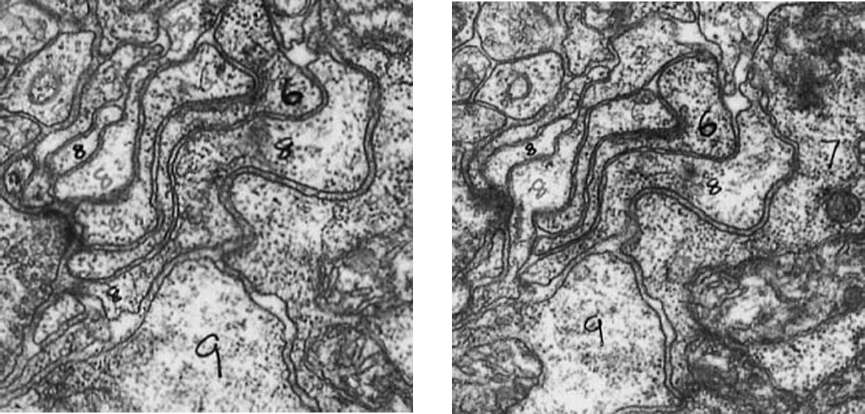
Figure 33. Tracing the branches of neurons by matching their cross-sections in successive slices
Â
In 1986 Brenner's team published the connectome as an entire issue of the
Philosophical Transactions of the Royal Society of London,
a journal of the same society that had welcomed Leeuwenhoek as a member centuries before. The paper was titled “The Structure of the Nervous System of the Nematode
Caenorhabditis elegans,
” but its running head was the pithier “The Mind of a Worm.” The body of the text is a 62-page appetizer. The main course is 277 pages of appendices, which describe the 302 neurons of the worm
along with their synaptic connections.
As Brenner had hoped, the
C. elegans
connectome turned out to be useful for understanding the neural basis of the worm's behaviors. For example, it helped identify the neural pathways important for behaviors like swimming away from a touch to the head.
But only a small fraction of Brenner's original ambitions were realized. It wasn't for lack of images; Nichol Thomson had gathered plenty of them, from many worms. He had actually imaged worms with many types of genetic defects, but it was too laborious to analyze the images to detect the hypothesized abnormalities in their connectomes. Brenner had started out wanting to investigate the hypothesis that the “minds” of worms differ because their connectomes differ, but he had been unable to do so because his team had found only a single connectome, that of a normal worm.
Finding even one connectome was by itself a monumental feat. Analyzing the images consumed over a dozen years of effort in the 1970s and 1980sâmuch more labor than was required to cut and image the slices. David Hall, another
C. elegans
pioneer, has made these images available online in a fascinating repository of information about the worm. (The vast majority of them remain unanalyzed today.) The toil of Brenner's team served as a cautionary note, effectively warning other scientists, “Don't try this at home.”
The situation began to improve in the 1990s, when computers became cheaper and more powerful. John Fiala and Kristen Harris
created a software program that facilitated the manual reconstruction of the shapes of neurons. The computer displayed images on a screen and allowed a human operator to draw lines on top of them using a mouse. This basic functionality, familiar to anyone who has used computers to create drawings, was then extended to allow a person to trace a neuron through a stack of images, drawing a boundary around each cross-section. As the operator worked, each image in the stack would become covered with many boundary drawings. The computer kept track of all the cross-section boundaries that belonged to each neuron, and displayed the results of the operator's labors by coloring within the lines. Each neuron was filled with a different color, so that the stack of images resembled a three-dimensional coloring book. The computer could also render parts
of neurites in three dimensions, as in the image shown in Figure 34.
Â
 Â
Â
Â
Figure 34. Three-dimensional rendering of neurite fragments reconstructed by hand
Â
With this process, scientists could do their work much more efficiently than Brenner's team had in the
C. elegans
project. Images were now stored neatly on the computer, so researchers no longer had to deal with thousands of photographic plates. And using a mouse was less cumbersome than manual marking with felt-tip pens. Nevertheless, analyzing the images still required human intelligence and was still extremely time-consuming. Using their software to reconstruct tiny pieces of the hippocampus and the neocortex, Kristen Harris and her colleagues discovered many interesting facts about axons and dendrites. The pieces were so small, however, that they contained only minuscule fragments of neurons. There was no way to use them to find connectomes.
Based on the experience of these researchers, we can extrapolate that manual reconstruction of just one cubic millimeter of cortex could take a million person-years,
much longer than it would take to collect the electron microscopic images. Because of these daunting numbers, it's clear that the future of connectomics hinges on automating image analysis.
Â
Ideally we'd have a computer, rather than a person, draw the boundaries of each neuron. Surprisingly, though, today's computers are not very good at detecting boundaries, even some that look completely obvious to us. In fact, computers are not so good at any visual task. Robots in science fiction movies routinely look around and recognize the objects in a scene, but researchers in artificial intelligence (AI) are still struggling to give computers even rudimentary visual powers.
In the 1960s researchers hooked up cameras to computers and attempted to build the first artificial vision systems. They tried to program a computer to turn an image into a line drawing, something any cartoonist could do. They figured it would be easy to recognize the objects in the drawing based on the shape of their boundaries. It was then that they realized how bad computers are at seeing edges. Even if the images were restricted only to stacks of children's blocks, it was challenging for the computers to detect the boundaries of the blocks.
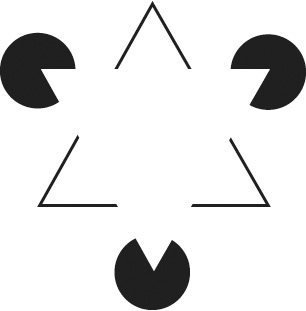
Why is this task so difficult for computers? Some subtleties of boundary detection are revealed by a well-known illusion called the Kanizsa triangle (Figure 35). Most people see a white triangle superimposed on a black-outlined triangle and three black circles. But it's arguable that the white triangle is illusory. If you look at one of its corners while blocking the rest of the image with your hand, you'll see a partially eaten pie (or a Pac-Man, if you remember that video game from the 1980s) rather than a black circle. If you look at one of the V's while blocking the rest of the image with both hands, you won't see any boundary where you used to see a side of the white triangle. That's because most of the length of each side is the same color as the background, with no jump in brightness. Your mind fills in the missing parts of the sidesâand perceives the superimposed triangleâonly when provided with the context of the other shapes.
Â
 Â
Â
Â
Figure 35. The “illusory contours” of the Kanizsa triangle
Â
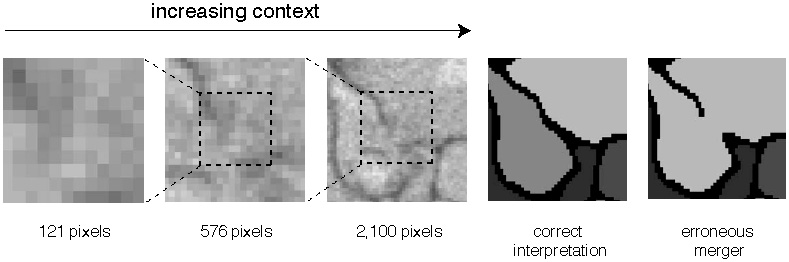
This illusion might seem too artificial to be important for normal vision. But even in images of real objects, context turns out to be essential for the accurate perception of boundaries. The first panel of Figure 36, a zoomed-in view of part of an electron microscope image of neurons, shows little evidence of a boundary. As subsequent panels reveal more of the surrounding pixels, a boundary at the center becomes evident. Detecting the boundary leads to the correct interpretation of the image (next-to-last panel); missing the boundary would lead to an erroneous merger of two neurites (last panel). This kind of mistake, called a merge error, is like a child's use of the same crayon to color two adjacent regions in a coloring book. A split error (not shown) is like the use of two different crayons to color a single region.
Â
 Â
Â
Â
Figure 36. The importance of context for boundary detection
Â
Granted, this sort of ambiguity is relatively rare. The one shown in the figure presumably arose because the stain failed to penetrate one location in the tissue. In most of the rest of the image, however, it would be obvious whether or not there is a boundary even in a zoomed-in view. Computers are able to detect boundaries accurately at these easy locations but still stumble at a few difficult ones, because they are less adept than humans at using contextual information.
Boundary detection is not the only visual task that computers need to perform better if we want to find connectomes. Another task involves recognition. Many digital cameras are now smart enough to locate and focus on the faces in a scene. But sometimes they erroneously focus on some object in the background, showing that they still don't recognize faces as well as people do. In connectomics, we'd like computers to perform a similar task, and to do it flawlessly: look through a set of images and find all the synapses.
Why have we failed (so far) to create computers that see as well as humans? In my view, it is because we see so well. The early AI researchers focused on duplicating capabilities that demand great effort from humans, such as playing chess or proving mathematical theorems. Surprisingly, these capabilities ended up being not so difficult for computersâin 1997 IBM's Deep Blue supercomputer defeated the world chess champion Garry Kasparov. Compared with chess, vision seems childishly simple: We open our eyes and instantly see the world around us. Perhaps because of this effortlessness, early AI researchers didn't anticipate that vision would be so difficult for machines.
Sometimes the people who are the best at doing something are the worst teachers. They themselves can do the task unconsciously, without thinking, and if they're asked to explain what they do, they have no idea. We are all virtuosos at vision. We've always been able to do it, and we can't understand an entity that can't. For these reasons we're lousy at teaching vision. Luckily we never have to, except when our students are computers.
In recent years some researchers have given up on instructing computers to see. Why not let a computer teach itself? Collect a huge number of examples of a visual task performed by humans, and then program a computer to imitate these examples. If the computer succeeds, it will have “learned” the task without any explicit instruction. This method is known as machine learning,
and it's an important subfield of computer science. It has yielded the digital cameras that focus on faces, as well as many other successes in AI.
Several laboratories around the world, including my own, are using machine learning to train computers to see neurons. We start by making use of the kind of software that John Fiala and Kristen Harris developed. People manually reconstruct the shapes of neurons, which serve as examples for the computer to emulate. Viren Jain and Srini Turaga,
my doctoral students at the time we began the work, have devised methods for numerically “grading” a computer's performance by measuring its disagreement with humans. The computer learns to see the shapes of neurons by optimizing its “grade” on the examples. Once trained in this way, the computer is given the task of analyzing images that humans have not manually reconstructed. Figure 37 shows a computer reconstruction of retinal neurons. This approach, though still in its beginning stages, has already attained unprecedented accuracy.
