Why Evolution Is True (13 page)
Read Why Evolution Is True Online
Authors: Jerry A. Coyne

And the evolutionary prediction that we’ll find pseudogenes has been fulfilled—amply. Virtually every species harbors dead genes, many of them still active in its relatives. This implies that those genes were also active in a common ancestor, and were killed off in some descendants but not in others.
17
Out of about thirty thousand genes, for example, we humans carry more than two thousand pseudogenes. Our genome—and that of other species—are truly well populated graveyards of dead genes.
17
Out of about thirty thousand genes, for example, we humans carry more than two thousand pseudogenes. Our genome—and that of other species—are truly well populated graveyards of dead genes.
The most famous human pseudogene is GLO, so called because in other species it produces an enzyme called L-gulono-γ-lactone oxidase. This enzyme is used in making vitamin C (ascorbic acid) from the simple sugar glucose. Vitamin C is essential for proper metabolism, and virtually all mammals have the pathway to make it—all, that is, except for primates, fruit bats, and guinea pigs. In these species, vitamin C is obtained directly from their food, and normal diets usually have enough. If we don’t ingest enough vitamin C, we get sick: scurvy was common among fruit-deprived seamen of the nineteenth century. The reason why primates and these few other mammals don’t make their own vitamin C is because they don’t need to. Yet DNA sequencing tells us that primates still carry most of the genetic information needed to make the vitamin.
It turns out that the pathway for making vitamin C from glucose involves a sequence of four steps, each promoted by the product of a different gene. Primates and guinea pigs still have active genes for the first three steps, but the last step, which requires the GLO enzyme, doesn’t take place: GLO has been inactivated by a mutation. It has become a pseudogene, called
yrGLO
(ψ is the Greek letter psi, standing for “pseudo”).
ψGLO
doesn’t work because a single nucleotide in the gene’s DNA sequence is missing. And it’s exactly the
same
nucleotide missing in other primates. This shows that the mutation that destroyed our ability to make vitamin C was present in the ancestor of all primates, and was passed on to its descendants. The inactivation of GLO in guinea pigs happened independently, since it involves different mutations. It’s highly likely that since fruit bats, guinea pigs, and primates got plenty of vitamin C in their diet, there was no penalty for inactivating the pathway that made it. This could even have been beneficial since it eliminated a protein that might have been costly to produce.
yrGLO
(ψ is the Greek letter psi, standing for “pseudo”).
ψGLO
doesn’t work because a single nucleotide in the gene’s DNA sequence is missing. And it’s exactly the
same
nucleotide missing in other primates. This shows that the mutation that destroyed our ability to make vitamin C was present in the ancestor of all primates, and was passed on to its descendants. The inactivation of GLO in guinea pigs happened independently, since it involves different mutations. It’s highly likely that since fruit bats, guinea pigs, and primates got plenty of vitamin C in their diet, there was no penalty for inactivating the pathway that made it. This could even have been beneficial since it eliminated a protein that might have been costly to produce.
A dead gene in one species that is active in its relatives is evidence for evolution, but there’s more. When you look at
ψGLO
in living primates, you find out that its sequence is more similar between close relatives than between more distant ones. The sequences of human and chimp ψ
GLO
, for example, resemble each other closely, but differ more from the
ψGLO
of orangutans, which are more distant relatives. What’s more, the sequence of guinea pig
ψGLO
is very different from that of all primates.
ψGLO
in living primates, you find out that its sequence is more similar between close relatives than between more distant ones. The sequences of human and chimp ψ
GLO
, for example, resemble each other closely, but differ more from the
ψGLO
of orangutans, which are more distant relatives. What’s more, the sequence of guinea pig
ψGLO
is very different from that of all primates.
Only evolution and common ancestry can explain these facts. All mammals inherited a functional copy of the GLO gene. About 40 million years ago, in the common ancestor of all primates, a gene that was no longer needed was inactivated by a mutation. All primates inherited that same mutation. After GLO was silenced, other mutations continued to occur in the gene that was no longer expressed. These mutations accumulated over time—they are harmless if they occur in genes that are already dead—and were passed on to descendant species. Since closer relatives share a common ancestor more recently, genes that change in a time-dependent way follow the pattern of common ancestry, leading to DNA sequences more similar in close than in distant relatives. This occurs whether or not a gene is dead. The sequence of
i/rGLO
in guinea pigs is so different because it was inactivated independently, in a lineage that had already diverged from that of primates. And
ψGLO
is not unique in showing such patterns: there are many other such pseudogenes.
i/rGLO
in guinea pigs is so different because it was inactivated independently, in a lineage that had already diverged from that of primates. And
ψGLO
is not unique in showing such patterns: there are many other such pseudogenes.
But if you believe that primates and guinea pigs were specially created, these facts don’t make sense. Why would a creator put a pathway for making vitamin C in all these species, and then inactivate it? Wouldn’t it be easier simply to omit the whole pathway from the beginning? Why would the same inactivating mutation be present in all primates, and a different one in guinea pigs? Why would the sequences of the dead gene exactly mirror the pattern of resemblance predicted from the known ancestry of these species? And why do humans have thousands of pseudogenes in the first place?
We also harbor dead genes that came from other species, namely viruses. Some, called “endogenous retroviruses,” can make copies of their genome and insert them into the DNA of species they infect. (HIV is a retrovirus.) If the viruses infect the cells that make sperm and eggs, they can be passed on to future generations. The human genome contains thousands of such viruses, nearly all of them rendered harmless by mutations. They are the remnants of ancient infections. But some of these remnants sit in exactly the same location on the chromosomes of humans and chimpanzees. These were surely viruses that infected our common ancestor and were passed on to both descendants. Since there is almost no chance of viruses inserting themselves independently at exactly the same spot in two species, this points strongly to common ancestry.
Another curious tale of dead genes involves our sense of smell, or rather our poor sense of smell, for humans are truly bad sniffers among land mammals. Nevertheless, we can still recognize more than ten thousand different odors. How can we accomplish such a feat? Until recently, this was a complete mystery. The answer lies in our DNA—in our many olfactory receptor (OR) genes.
The OR story was worked out by Linda Buck and Richard Axel, who were awarded the Nobel Prize for this feat in 2004. Let’s look at OR genes in a super-sniffer: the mouse.
Mice depend heavily on their sense of smell, not only to find food and avoid predators, but also to detect one another’s pheromones. The sensory world of a mouse is vastly different from ours, in which vision is far more important than smell. Mice have about a thousand active OR genes. All of them descend from a single ancestral gene that arose millions of years ago and became duplicated many times, so that each gene differs slightly from the others. And each produces a different protein—an “olfactory receptor”—that recognizes a different airborne molecule. Each OR protein is expressed in a different type of receptor cell in the tissues lining the nose. Different odors contain different combinations of molecules, and each combination stimulates a different group of cells. The cells send signals to the brain, which integrates and decodes the different signals. That’s how mice can distinguish the smell of cats from that of cheese. By integrating
combinations
of signals, mice (and other mammals) can recognize far more odors than they have OR genes.
combinations
of signals, mice (and other mammals) can recognize far more odors than they have OR genes.
The ability to recognize different smells is useful: it enables you to distinguish kin from nonkin, find a mate, locate food, recognize predators, and see who’s been invading your territory. The survival advantages are enormous. How has natural selection tapped them? First, an ancestral gene became duplicated a number of times. Such duplication happens from time to time as an accident during cell division. Gradually, the duplicated copies diverged from each other, with each binding to a different odor molecule. A different type of cell evolved for each of the thousand OR genes. And at the same time, the brain became rewired to combine the signals from the various kinds of cells to create the sensations of different odors. This is a truly staggering feat of evolution, driven by the sheer survival value of the discerning sniff!
Our own sense of smell comes nowhere close to that of mice. One reason is that we express fewer OR genes—only about four hundred. But we still carry a total of eight hundred OR genes, which make up nearly 3 percent of our entire genome. And fully half of these are pseudogenes, permanently inactivated by mutations. The same is true for most other primates. How did this happen? Probably because we primates, who are active during the day, rely more on vision than on smell, and so don’t need to discriminate among so many odors. Unneeded genes eventually get bumped off by mutations. Predictably, primates with color vision, and hence greater discrimination of the environment, have more dead OR genes.
If you look at the sequences of human OR genes, both active and inactive, they are most similar to those of other primates, less similar to those of “primitive” mammals like the platypus, and less similar yet to the OR genes of distant relatives like reptiles. Why should dead genes show such a relationship, if not for evolution? And the fact that we harbor so many inactive genes is even more evidence for evolution: we carry this genetic baggage because it was needed in our distant ancestors who relied for survival on a keen sense of smell.
But the most striking example of the evolution-or de-evolution—of OR genes is the dolphin. Dolphins don’t need to detect volatile odors in the air, since they do their business underwater, and they have a completely different set of genes for detecting waterborne chemicals. As one might predict, OR genes of dolphins are inactivated. In fact, 80
percent of them
are inactivated. Hundreds of them still sit silently in the dolphin genome, mute testimony of evolution. And if you look at the DNA sequences of these dead dolphin genes, you’ll find that they resemble those of land mammals. This makes sense when we realize that dolphins evolved from land mammals whose OR genes became useless when they took to the water.
18
This makes no sense if dolphins were specially created.
percent of them
are inactivated. Hundreds of them still sit silently in the dolphin genome, mute testimony of evolution. And if you look at the DNA sequences of these dead dolphin genes, you’ll find that they resemble those of land mammals. This makes sense when we realize that dolphins evolved from land mammals whose OR genes became useless when they took to the water.
18
This makes no sense if dolphins were specially created.
Vestigial genes can go hand in hand with vestigial structures. We mammals evolved from reptilian ancestors that laid eggs. With the exceptions of the “monotremes” (the order of mammals that includes the Australian spiny anteater and duck-billed platypus), mammals have dispensed with egg-laying, and mothers nourish their young directly through the placenta instead of by providing a storehouse of yolk. And mammals carry three genes that, in reptiles and birds, produce the nutritious protein vitellogenin, which fills the yolk sac. But in virtually all mammals these genes are dead, totally inactivated by mutations. Only the egg-laying monotremes still produce vitellogenin, having one active and two dead genes. What’s more, mammals like ourselves still produce a yolk sac—but one that is vestigial and yolkless, a large, fluid-filled balloon attached to the fetal gut (figure 15). In the second month of human pregnancy, it detaches from the embryo.
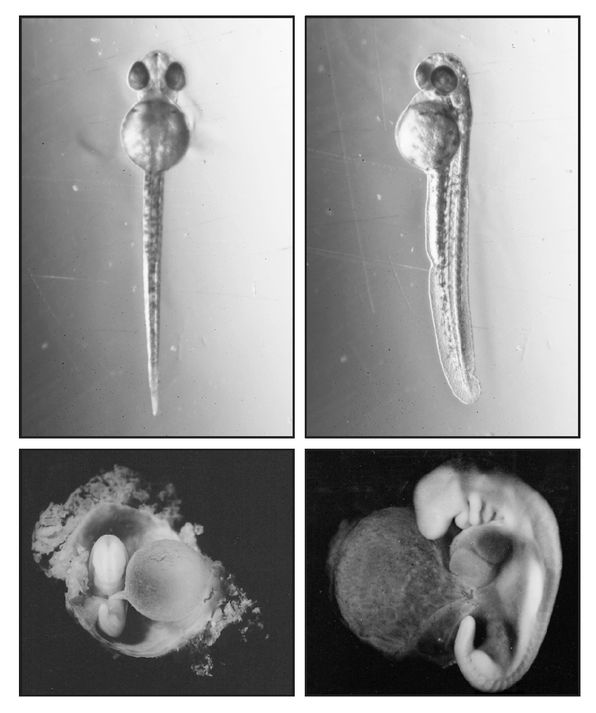
FIGURE 15.
Normal and vestigial yolk sacs. Top photos: full yolk sac of the embryonic zebra-fish,
Danio
rerio, extracted from the egg case at two days, just before hatching. Bottom photos: empty vestigial yolk sac of a human embryo at about four weeks. The human embryo at bottom right shows the branchial arches, the hindlimb bud, and the “tail” below the hindlimb.
Normal and vestigial yolk sacs. Top photos: full yolk sac of the embryonic zebra-fish,
Danio
rerio, extracted from the egg case at two days, just before hatching. Bottom photos: empty vestigial yolk sac of a human embryo at about four weeks. The human embryo at bottom right shows the branchial arches, the hindlimb bud, and the “tail” below the hindlimb.
With its ducklike bill, fat tail, poison-tipped spurs on the hind legs of males, and the ability of females to lay eggs, the platypus of Australia is bizarre in many ways. If ever a creature seems
un
intelligently designed—or perhaps devised for a creator’s amusement—it would be this one. But the platypus has one more odd feature: it lacks a stomach. Unlike nearly all vertebrates, who have a pouchlike stomach in which digestive enzymes break down food, the platypus “stomach” is just a slight swelling of the esophagus where it joins the intestine. This stomach completely lacks the glands that produce digestive enzymes in other vertebrates. We’re not sure why evolution has eliminated the stomach—perhaps the platypus diet of soft insects doesn’t require much processing—but we know that the platypus came from ancestors with stomachs. One reason is that the platypus genome contains two pseudogenes for enzymes related to digestion. No longer needed, they’ve become inactivated by mutation, but still testify to the evolution of this strange beast.
Palimpsests in Embryosun
intelligently designed—or perhaps devised for a creator’s amusement—it would be this one. But the platypus has one more odd feature: it lacks a stomach. Unlike nearly all vertebrates, who have a pouchlike stomach in which digestive enzymes break down food, the platypus “stomach” is just a slight swelling of the esophagus where it joins the intestine. This stomach completely lacks the glands that produce digestive enzymes in other vertebrates. We’re not sure why evolution has eliminated the stomach—perhaps the platypus diet of soft insects doesn’t require much processing—but we know that the platypus came from ancestors with stomachs. One reason is that the platypus genome contains two pseudogenes for enzymes related to digestion. No longer needed, they’ve become inactivated by mutation, but still testify to the evolution of this strange beast.
WELL BEFORE THE TIME OF DARWIN, biologists were busy studying both embryology (how an animal develops) and comparative anatomy (the similarities and differences in the structure of different animals). Their work turned up many peculiarities that, at the time, didn’t make sense. For example, all vertebrates begin development in the same way, looking rather like an embryonic fish. As development proceeds, different species begin to diverge—but in weird ways. Some blood vessels, nerves, and organs that were present in the embryos of all species at the start suddenly disappear, while others go through strange contortions and migrations. Eventually, the dance of development culminates in the very different adult forms of fish, reptiles, birds, amphibians, and mammals. Nevertheless, when development begins they look very much alike. Darwin tells the story of how the great German embryologist Karl Ernst von Baer became confused by the similarity of vertebrate embryos. Von Baer wrote to Darwin:
In my possession are two little embryos in spirit [alcohol], whose names I have omitted to attach, and at present I am quite unable to say to what class they belong. They may be lizards or small birds, or very young mammalia, so complete is the similarity in the mode of formation of the head and trunk in these animals.
Other books
Carnival of Death by Keene, Day
Creole Belle by Burke, James Lee
Cassie by Barry Jonsberg
Furever After by Arielle Lewenhart
Highlander Mine by Miller, Juliette
A World Alone (Dead World Series Book 1) by R.K. Weir
Hostile Takeover by Hill, Joey W
When the Devil Doesn't Show: A Mystery by Christine Barber
Jigsaw by Sybille Bedford
Play With Me by Alisha Rai
