Neanderthal Man (18 page)

Since we had struggled so long to retrieve nuclear DNA, Alex’s mammoth results were very welcome indeed and for several days I was very happy about them. But of course I wasn’t all that interested in mammoths. I was interested in Neanderthals, and I was painfully aware that there were no Neanderthals in permafrost. I urged Alex to go back and try the cave-bear remains from Vindija again, to see if he could retrieve nuclear DNA from remains that were not frozen. He analyzed mitochondrial DNA from several Croatian cave bears and identified one bone that seemed to contain a lot of it. We carbon-dated it and found it to be 33,000 years old, and thus roughly contemporaneous with the Neanderthals. Alex concentrated on this bone. He tried the ribosomal RNA gene that occurs in many copies in the genome. He obtained small amounts of an amplification product. Reconstructing its sequence from clones, he found that the sequence was indeed identical to that in present-day bears.
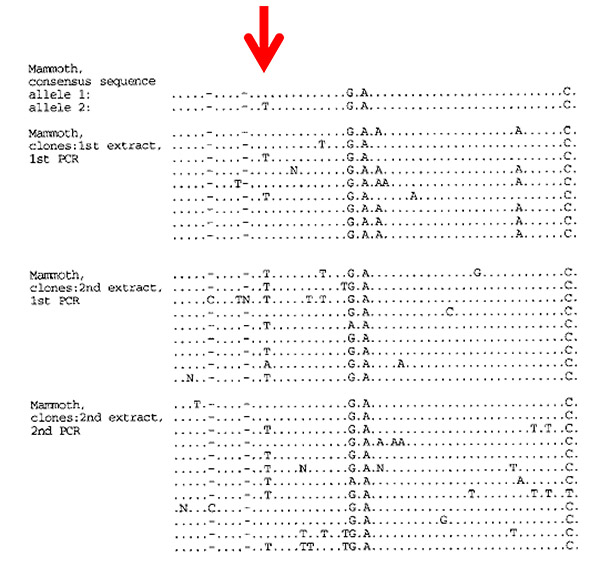
Figure 9.1. Cloned DNA sequences from three amplifications of a nuclear gene fragment from a 14,000-year-old mammoth. The arrow points to the first heterozygous position or SNP ever observed from the Late Pleistocene. From A. D. Greenwood et al., “Nuclear DNA sequences from Late Pleistocene megafauna,”
Molecular Biology and Evolution
16, 1466–1473 (1999).
This was a success, but one with a dark side. It was so hard to amplify this multicopy-gene fragment that any attempt to get single-copy genes such as the vWF gene he studied in the mammoth seemed doomed to fail. Alex tried it anyway, of course, but as expected he was unsuccessful. So, after all the excitement over the mammoth results, I was—secretly—deeply disappointed by these experiments. We had demonstrated that nuclear DNA could survive over tens of thousands of years in permafrost, but only traces of very common nuclear DNA sequences could be found in the bones of cave bears. There was an enormous difference between permafrost and limestone caves.
In 1999, we published Alex’s findings in what I considered to be a beautiful paper, though it was subsequently largely overlooked.
{40}
It demonstrated that nuclear DNA survives in remains found in permafrost and that even heterozygous positions, where the two chromosomes in an individual differ in DNA sequence, can be determined. We were optimistic about the prospects for genetic research in the permafrost, and we noted at the end of the paper that
A plethora of faunal remains exist in permafrost deposits and other cold environments. The fact that such remains can yield not only mtDNA but also single-copy nuclear DNA sequences in a substantial amount of cases opens up the possibilities of using nuclear loci in phylogenetic and population genetic studies and of studying genes determining phenotypic traits.
Eventually others would take up this line of work, although not for another five to ten years. Worse, barring the discovery of a Neanderthal in permafrost, it seemed we might never see the whole genome of a Neanderthal.
Going Nuclear
____________________
In the lab, I went about the business of supervising the experiments that pushed the work slowly but reliably forward. But whenever I was confined to a small seat during a long airplane ride, or to a darkened lecture hall during a seemingly irrelevant presentation at a conference, I came back to my greatest frustration: our inability to retrieve nuclear DNA from Neanderthals. I felt that it
had
to be there, even if the PCR could not retrieve it. We simply had to come up with a better way to find it.
A fresh attempt in this direction was made by Hendrik Poinar. Worn down by his fruitless quest for DNA from fauna and flora encased millions of years ago in amber, he had decided to move on to more promising endeavors. Fortunately for us I had just spent time at some boring conference lectures, where I got to thinking about the work we’d done on retrieving DNA from animal droppings. One of those we’d studied was the extinct American ground sloth, an Ice Age animal. The giant sloths had left behind large amounts of droppings, which archaeologists dressed up with the fancy name of coprolites. In fact, in some caves in places like Nevada, the entire floor, to some depth, is largely made up of old ground-sloth feces. In a paper in
Science
in 1998, Hendrik had already shown that mitochondrial DNA was preserved in such material and we described the retrieval of plant DNA from a single sloth bolus, showing how it could be used to reconstruct the ingredients of the meal the sloth had ingested shortly before its death 20,000 years ago.
{41}
This success suggested that lots of DNA, even nuclear DNA, was preserved in ancient fecal remains. I suggested that Hendrik try to find it.
Hendrik had begun the search with a chemical trick we had developed the year before. Back in 1985, when I was analyzing the mummies from Berlin, I had noticed that almost all of the extracts contained a component that produced blue fluorescence in UV light, and that if an extract fluoresced blue, it never yielded DNA. I did not know what this component was, but the observation was painfully memorable because of the disappointment I felt when I saw the blue instead of the pink glow I was hoping for. As I learned more about the chemistry that may have gone on in dead tissues over thousands of years, I came across the phenomenon known as the Maillard reaction, a chemical reaction much studied by the food industry. As it happened, my mother was a food chemist, so she sent me lots of literature on it. The Maillard reaction occurs when common forms of sugar are heated or persist at less-hot temperatures for a long time. They then form chemical cross-links with amino groups found in proteins and in DNA, resulting in large, tangled molecular complexes. The Maillard reaction occurs in many forms of cooking, and side products of the Maillard reaction result in the pleasant smell and color of freshly baked bread. But most interesting to me was that Maillard products give off a blue fluorescence in UV light. I thought that this might have been what was going on with the Egyptian mummies. I associated this reaction not only with the blue fluorescence of the mummy extracts but also (perhaps incorrectly) with their brown color and their characteristic smell, which is sweet and not unpleasant. And I wondered if the reason I couldn’t extract DNA from them was that the Maillard reaction bonded it to other molecules.
There was a way to find out. In 1996, a paper in
Nature
described a chemical reagent—N-phenacylthiazolium bromide, abbreviated PTB—that could break down the complexes formed by the Maillard reaction.
{42}
PTB, when added to baked bread, would turn it into dough again (albeit, surely, not dough that anyone would be tempted to put back in the oven). Since PTB could not be bought commercially, Hendrik synthesized it in the lab. When we added PTB to extracts from ancient samples of cave bears and Neanderthals, it indeed sometimes resulted in better amplification. And when Hendrik added PTB to extracts from the 20,000-year-old Nevada coprolites, he was able to amplify fragments of the vWF gene that Alex had partially sequenced from mammoths as well as fragments from two other nuclear genes, all to my great surprise. We published this work in July 2003,
{43}
finally demonstrating that the nuclear genome could be preserved even when remains were not frozen.
Encouraged by those results, I felt that there was now every reason to persist in our attempts to retrieve nuclear DNA from the bones of cave bears—again, with PTB. But sadly, this time the chemical trick didn’t help. Actually, it turned out that the Nevada coprolites were a rare exception where PTB could turn failure into success. The coprolites did, however, confirm my feeling that the nuclear DNA was there, and that we simply needed new techniques to find it.
To get ideas about new techniques I took to consulting as many people as possible about ways to sequence small amounts of DNA. One of the people subjected to my questioning was the Swedish biochemist Mathias Uhlén, a creative inventor and biotech entrepreneur. Mathias combines an apparently unlimited energy and a childlike enthusiasm for new ideas with a knack for collecting creative people around him and transmitting his enthusiasm to them. I always felt energized after my encounters with him. One of the many creative people around Mathias was Pål Nyrén. Ten years earlier, he had conceived of a new technique for DNA sequencing he had developed in spite of widespread skepticism. Mathias had realized the potential of Pål’s idea. He also saw that it was timely to think about new ways to sequence DNA: we were still using the approach invented by Fred Sanger in the UK, which, in 1980, had earned him his second Nobel Prize in chemistry.
The Sanger sequencing method relies on the sequential incorporation of the four nucleotides by a DNA polymerase, an enzyme that makes new DNA strands using old ones as templates. In such a sequencing reaction, the DNA polymerase starts its synthesis of DNA strands from a primer at a defined point in the DNA. A small fraction of each of the four nucleotides are labeled with different fluorescent dyes and chemically modified so that when the DNA polymerase builds them in, the synthesis stops. This process creates DNA strands of different lengths, each with the dye at its end indicating which nucleotide sits there. The fragments thus terminated and labeled can be separated by electrophoresis in a gel according to their size. This reveals which dye and thus nucleotide is present—for example, ten positions away from where the synthesis started, eleven positions away, twelve positions away, and so on. The best machines employed to sequence DNA—for example, by the Human Genome Project—can sequence almost a hundred pieces of DNA at a time, for stretches as long as 800 nucleotides. What Pål developed in Mathias’s lab was a method called pyrosequencing. Though still in its infancy, this method was potentially much faster and simpler than Sanger sequencing.
Pyrosequencing also uses a DNA polymerase to build DNA sequences, but it detects each nucleotide incorporated into the DNA not by cumbersome separation of fragments according to size but by a flash of light emitted after each nucleotide is built into the DNA chain. The trick Pål had devised was to add just one of the four nucleotides at a time to the reaction mix. For example, if an A (adenine) is added, and the strand that is being used as a template at that point carries a T (thymine, which pairs with adenine), the DNA polymerase builds the A into the growing strand, and an enzymatic system in the reaction causes a light signal to be generated. This flash of light is detected by a powerful camera and registered by a computer. If the template strand carries not a T but another base, no flash of light is generated. Pål added each of the four nucleotides consecutively, cycle after cycle. By noting the light flashes, he could read the order of nucleotides in a DNA fragment. It was a brilliant method, relying only on pumping the nucleotides and other reagents into a reaction chamber and taking pictures with a camera. Even more importantly, it could easily be automated. When Mathias told me about it, I became as enthusiastic as he was.
A little later, Mathias asked me to serve on the scientific advisory board of a company called Pyrosequencing, which he and Pål had founded to produce a commercial instrument for performing this technique. I gladly agreed, since doing so would give me an opportunity to keep up with the development of an exciting technology that I thought might transform how we studied ancient DNA. I joined the advisory board in 2000, a year after the company had produced its first commercial instrument, which could simultaneously sequence ninety-six different DNA fragments, each isolated in a well on a plastic plate. However, from each fragment it could read only about thirty consecutive nucleotides. This was deeply unimpressive compared with contemporary machines relying on the Sanger principle, but pyrosequencing was a young technology and had not reached the limits of its possibilities. In fact, although I did not fully appreciate this at the time, it represented the beginning of a revolution, known as “second-generation sequencing,” that would fundamentally change not just our investigations of ancient DNA but many aspects of biology.
I very much wanted to try out pyrosequencing, so I asked Henrik Kaessmann to spend some time in Mathias’s lab at the Royal Institute of Technology in Stockholm. Henrik welcomed the opportunity to surprise people in Stockholm with his flawless Swedish; although he grew up in southern Germany, he speaks the language fluently thanks to his Swedish mother. He was also able to generate data from present-day human populations in Europe and Asia that would help in showing how they were related to one another. As with all new techniques, this required learning new skills and some troubleshooting, but it worked well.
In August 2003, the board of Pyrosequencing decided to license the technology to 454 Life Sciences, a US company founded by the biotech entrepreneur Jonathan Rothberg. 454 Life Sciences intended to enhance pyrosequencing with state-of-the-art fluidics. Its innovation relied on adding short synthetic pieces of DNA to the ends of DNA molecules. Single strands of DNA were then captured on beads and ingeniously amplified in little oil bubbles, allowing hundreds of thousands of different strands to be amplified separately but simultaneously in one big reaction. Then the beads were separated from one another on a plate with hundreds and thousands of wells for the pyrosequencing step. Finally (and crucially), to keep tabs on which wells were emitting flashes of light from cycle to cycle, the company used image-tracking methods borrowed from astronomers, who track millions of stars in the night sky. This allowed it to simultaneously sequence not ninety-six but two hundred thousand DNA fragments at a time!
