Ancient DNA: Methods and Protocols (20 page)
Read Ancient DNA: Methods and Protocols Online
Authors: Beth Shapiro

Key words:
Ancient DNA , Chimpanzees , DNA damage , Genetic diversity , Mitochondrial DNA (mtDNA) ,
Museum collections
,
Nondestructive DNA extraction
,
Phylogeography ,
Population
extinction
*
Note
: In the case study presented in this chapter, we describe DNA extraction and amplifi cation of mitochondrial DNA from historic chimpanzee samples from museum collections using a method similar to that presented in Chapter 13 . We discuss specifi c challenges associated with nondestructive DNA extraction, including contamination and DNA damage.
Beth Shapiro and Michael Hofreiter (eds.),
Ancient DNA: Methods and Protocols
, Methods in Molecular Biology, vol. 840, DOI 10.1007/978-1-61779-516-9_14, © Springer Science+Business Media, LLC 2012
101
102
E. Mohandesan
et al.
1. Introduction
Museum specimens represent one of the major sources of ancient DNA. Museum collections are valuable because they often contain rare or extinct species as well as large numbers of conspecifi c specimens that can be used to reveal the biological history of species and populations. Methods for DNA extraction from bones, teeth, and skin ar
e well established ( 1, 2
) . However, for almost all of these, a piece of tooth, bone, or skin has to be removed and dissolved prior to DNA extraction.
To circumvent this limitation, a nondestructive DNA extraction method has been developed, with a reported success rate of 90% for bones up to 164 years old
( 3
) . The protocol, described in detail in Chapter 13 , involves soaking the sample in GuSCN buffer and subsequently processing the buffer. Because it does not require the removal of a large piece of the specimen, this method prevents signifi cant damage to the specimen, leaving it intact for future analyses. In addition, if necessary, the DNA extraction can be repeated 3–5 times without signifi cant damage occurring to the specimen
( 3
) .
Here, we apply this nondestructive DNA extraction method to a large number of museum-preserved chimpanzee specimens. We discuss the success rate of this method, problems that arise during the procedure, and phylogenetic analyses performed subsequent to extraction and sequencing.
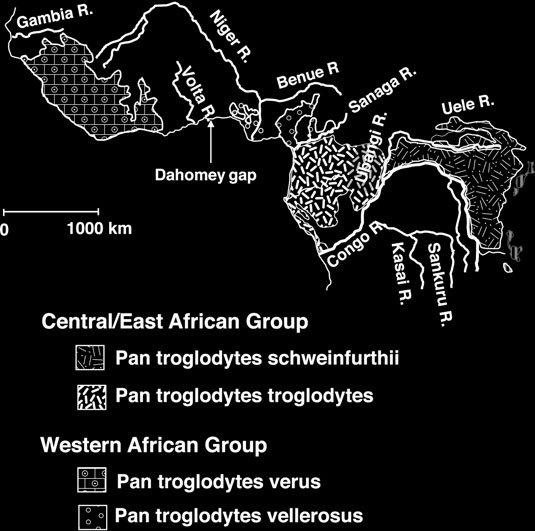
Common chimpanzees (
Pan troglodytes
) are traditionally
divided into three populations or subspecies based on geographic barriers (mostly rivers): west African
P. t. verus
( 4 )
, central African P.t. troglodytes , and east African
P. t. schweinfurthii
( 5, 6
)
.
Additional sampling in northern Cameroon/southern Nigeria has led to the designation of a fourth chimpanzee subspecies,
P. t. vellerosus
( 7– 11
) , although the phylogenetic distinctiveness and therefore the validity of this fourth chimpanzee subspecies is still debated
( 12
) . A recent analysis of about 300 microsatellites demonstrated convincingly that low levels of gene fl ow are occurring among the thr
ee traditionally accepted chimpanzee subspecies
However, due to a lack of captive individuals of
P. t. vellerosus
, the status of this potential subspecies has yet to be ascertained
( 12
) . Because chimpanzee populations have declined severely during the last decades
( 13– 15 )
, accessing genetic material from historic chimpanzee specimens should allow a better understanding of the geographical distribution and the population history of chimpanzees.
14 Case Study: Using a Nondestructive DNA Extraction Method…
103
2. Materials
and Methods
2.1. Sample
We used two rooms during the experiment so that sample prepara-
Preparation
tion could be kept separate from contamination-susceptible steps including buffer preparation and PCR setup. In the second room, we carried out buffer preparation and setup of PCR reagent mix in one fume hood, and DNA extraction and the addition of DNA
extract to the PCR in a second hood. In order to prevent modern DNA from potentially contaminating the experiments, we washed all working surfaces with 10–13% sodium hypochlorite solution (bleach) prior to DNA extraction. Both rooms were designated for ancient DNA work, and were spatially separated from all laboratories in which work on modern DNA was performed. The ancient DNA clean rooms were further isolated from any other area by an ante-room, which was used for decontaminating consumables and changing clothes.
We collected teeth from 86 chimpanzee (
Pan troglodytes
) individuals originating from different geographical locations in Africa and that are currently held in different museum collections. The fi nal data set comprised specimens from 35 eastern, 20 central, two western and one western/central (Nigeria-Cameroon) locations.
2.2. DNA Extraction
Prior to extraction, we prepared TE buffer, extraction solution,
and Amplifi cation
binding buffer, washing buffer, and silica suspension as described in Chapter 13 . We designed two overlapping primer pairs (A and B; see T
able 1 ) using Primer 3 version 0.4.0 (
http://frodo.wi.mit.
edu/primer3/ ). The primers were synthesized in 100 m M stock concentration and stored at −20ºC. For use in PCR, we diluted the primers to 10 m M concentration with HPLC-grade water and stored them at −20ºC.
Table 1
Primer designed for amplifying the investigated D-loop region of chimpanzee mtDNA
Primer sequence 5–3
¢
Product size
Primer pair A
Outer sense2 (OS2)
5 ¢ -CGC TAT GTA TTT CGT ACA TTA CT-3 ¢
210 bp
Inner antisense3 (IAS3)
5 ¢ -RTA GGT TTG TTG ATA TYR G-3 ¢
Primer pair B
Inner sense3 (IS3)
5 ¢ -TCA ACT CTC AAC TRT CRM ACA TA-3 ¢
130 bp
Outer antisense2 (OAS2)
5 ¢ -GAT TTG ACT GTA ATG TGC TAT G-3 ¢
104
E. Mohandesan
et al.
For extraction, we fi rst cleaned the surface of each specimen using a tissue moistened with HPLC-grade water. Removing dirt from the surface of the samples reduces the amount of substances that might inhibit the DNA extraction and/or the following enzymatic manipulations of DNA extract such as PCR.
We then soaked the samples in 5 mL extraction solution (L6
buffer) and incubated them at room temperature in the dark with constant slow rotation. After 5–7 days, we removed the buffer and rinsed the sample with HPLC-grade water. We dried the samples at room temperature in preparation for return to the museums from which they were obtained.
To continue with the DNA extraction, we transferred the buffer into a new 15-mL centrifuge tube. We added 50–100 m L of silica suspension (after vortexing the silica suspension to be certain that it was adequately mixed) and incubated the mixture for 1–3 h at room temperature with rotation. We then centrifuged the buffer at 1,800 ×
g
for 2 min and either discarded the supernatant or stored it at 4°C for later use. Next, we washed the silica pellet with 1 mL L2 buffer by pipetting up and down. We transferred the resuspended mixture to a 2-mL Eppendorf tube. This transfer makes handling more convenient, as 2-mL tubes rather than 15-mL
tubes can be used in all the following steps. We pelleted the silica via centrifugation for 5 s at 16,000 ×
g
, discarded the supernatant, and carefully removed any remaining liquid using a 200-m L pipette.
If the binding solution (L2 buffer) is not completely removed in this step, the salt concentration in the elution buffer will be too high, thus preventing the DNA from being completely released from the silica during elution.
We then washed the pellet with 1 mL washing buffer by
pipetting up and down. We centrifuged the resuspended mixture for 10 s at 16,000 ×
g
. We discarded the supernatant and removed the remaining liquid again carefully with a pipette. We dried the pellets at 56°C for 5 min or approximately 15 min at room temperature with open lids. We then added 100 m L TE (1×) to the pellet, incubated the mixture for 8 min at 65°C, and resuspended the pellet by stirring with the pipette tip and pipetting up and down. Finally, we centrifuged the eluate at 16,000 ×
g
for 1 min and transferred the supernatant into a new 2-mL Eppendorf tube, being careful not to leave any trace of silica. For some specimens, second and third extractions starting at the incubation step were subsequently perfor
med (see Subheading 3
).
We used the obtained extracts to generate an approximately 225 bp fragment of the HVR-I region of chimpanzee mtDNA by PCR amplifying two overlapping fragments of 210 and 130 bp, respectively, using primer pairs A (OS2/IAS3) and B (IS3/OAS2; see T
able 1
). PCR was carried out in 20 m L volumes containing 1×
PCR buffer (Applied Biosystems), 4 mM MgCl (Applied
2
Biosystems), 1 mg/mL BSA (Invitrogen), 0.5 mM mixed dNTPs
14 Case Study: Using a Nondestructive DNA Extraction Method…
105
(in equal concentrations; Amersham Biosciences), 0.25 m M of each primer (MWG-Biotech AG), 0.5–1 U of Taq Gold DNA polymerase (Applied Biosystems), and 5 m L DNA template (irrespective of DNA concentration). The initial denaturation step (94°C
for 4 min) was followed by 60 cycles of denaturation at 93°C for 20 s, binding of primers at 51°C (primer pair A) and 53°C (primer pair B) for 30 s and strand replication at 72°C for 30 s, followed by a fi nal extension at 72°C for 10 min. The PCR products were subjected to electrophoresis in 1.5% agarose, stained with ethidium bromide (50 ng/mL) and visualized over UV light. We included one negative control for every seven PCR reactions. Each fragment was amplifi ed twice for each specimen.
We purifi ed PCR products of the expected length with the
QIAquick Gel Extraction Kit (QIAGEN, Germany), and cloned
them using the TOPO TA
® Cloning Kit (Invitrogen, The
Netherlands) according to the manufacturer’s instruction. We sequenced the insert sequences for eight clones per sample on an ABI 3700 capillary sequencer after colony PCR and purifi cation on a QIAGEN BioRobot 9600.
2.3. Phylogenetic
We aligned the nucleotide sequences from the HVR-I regions
Analysis
sequenced from 56 chimpanzees in BioEdit version 7.0
( 16
) using CLUSTAL-W software. We checked the authenticity of obtained DNA sequences using BlastSearch (National Center for
Biotechnology Information)
( 17
) and reconstructed the phylogenetic relationship between the recovered sequences as well as extant chimpanzee sequences obtained from GenBank by constructing a serial network
( 18 )
. The serial network was created using the open-source R script TempNet (available at www.stanford.edu/group/
hadlylab/tempnet/ ). TempNet uses statistical parsimony to illustrate within-species relationships through time.
3. Results
and Discussion
Using the silica-based nondestructive method, we successfully amplifi ed and sequenced mtDNA sequences from 65% (56 of 86) of the chimpanzee specimens that were stored in different museums.
Of these, 53 samples (95%) yielded both PCR products, while the remaining three samples (5%) could only be partially amplifi ed.
All recovered sequences showed between 98 and 100% BLAST
similarity to chimpanzee mtDNA sequences archived in GenBank.
Analysis of consensus and clone sequences generated from two independent PCRs revealed identical sequences for 29 museum specimens (apart from C to T changes in individual clones, which are almost certainly due to DNA damage; see below) and multiple sequence variants within the remaining 26 (one sample could only 106
E. Mohandesan
et al.
be amplifi ed once and was excluded from further analyses). Thus, just over half of the samples yielded identical sequences across multiple PCRs, although for six of the samples yielding additional sequences, these occurred at such a low frequency that a likely endogenous sequence could be inferred. This overall result most likely indicates that cross-contamination occurred between museums specimens, especially since the sequence variants recovered sometimes belong to different chimpanzee subspecies. To investigate this further, we performed additional nondestructive extractions on 16 of the specimens that had yielded ambiguous sequences.
This additional experiment was motivated by the realization that the fi rst extraction may recover not only endogenous DNA but also any potential surface contaminant DNA, including cross-contamination that may have occurred as researchers handled multiple specimens. Additional extractions performed after the fi rst extraction should therefore be less likely to recover surface contaminants.
We performed second and in some cases third DNA extractions from 16 of the samples with variant sequences. Each extraction yielded less amplifi able DNA than the previous extraction, as judged by the number of failing PCRs and the strength of the product when amplifi cations were successful. However, the amount of DNA contamination was also reduced to some extent, and a likely endogenous DNA sequence could eventually be deduced for 9 of these 16
samples, while the remaining seven samples could not be resolved.
Thus, in total we were able to recover reproducible sequences from 44 samples, resulting in a total of 39 distinct haplotypes.
This result is in stark contrast to previous experience with this protocol when no evidence for contamination was obser
ved ( 3,
19, 20
) . However, while it should be noted that two of these previous studies were performed on small mammal specimens, where both storage conditions and, due to the fragile nature of the specimens, extraction kinetics might be different, the initial study introducing this method used both chimpanzee and hyena teeth. It is not clear why the results of this study differ so much from those of previous studies. One potential cause may lie in differences in museum storage and handling conditions that might have facilitated cross-contamination among the samples used in this study, but it is impossible to ascertain this possibility. Another fact worth mentioning is a high incidence of C to T changes, indicative of
DNA damage ( 21
) in our results. Thus, of the 29 samples that yielded unambiguous sequences, 26 showed C to T changes in individual clones. This observation suggests not only high DNA damage but also low DNA concentrations in these samples, making them more susceptible to contamination. Independent of the eventual cause for the high contamination rate on the samples used, our results show that studies on museum specimens face similar problems as those using fossil DNA, at least when using this extraction method. Therefore, similar precautions such as multiple